56. Bayesian versus Frequentist Decision Rules#
Contents
import matplotlib.pyplot as plt
import numpy as np
from numba import jit, prange, float64, int64
from numba.experimental import jitclass
from math import gamma
from scipy.optimize import minimize
56.1. Overview#
This lecture follows up on ideas presented in the following lectures:
A Problem that Stumped Milton Friedman described a problem that a Navy Captain presented to Milton Friedman during World War II.
The Navy had told the Captain to use a decision rule for quality control.
In particular, the Navy had ordered the Captain to use an instance of a frequentist decision rule.
The Captain doubted that that rule was a good one.
Milton Friedman recognized the Captain’s conjecture as posing a challenging statistical problem that he and other members of the US Government’s Statistical Research Group at Columbia University proceeded to try to solve.
A member of the group, the great mathematician and economist Abraham Wald, soon solved the problem.
A good way to formulate the problem is to use some ideas from Bayesian statistics that we describe in this lecture Exchangeability and Bayesian Updating and in this lecture Likelihood Ratio Processes, which describes the link between Bayesian updating and likelihood ratio processes.
The present lecture uses Python to generate simulations that evaluate expected losses under frequentist and Bayesian decision rules for an instance of the Navy Captain’s decision problem.
The simulations confirm the Navy Captain’s hunch that there is a better rule than the one the Navy had ordered him to use.
56.2. Setup#
To formalize the problem that had confronted the Navy Captain, we consider a setting with the following parts.
Each period a decision maker draws a non-negative random variable
The decision maker observes a sample
The decision maker wants to decide which distribution actually governs
He is worried about two types of errors and the losses that they will impose on him.
a loss
a loss
The decision maker pays a cost
We mainly borrow parameters from the quantecon lecture
A Problem that Stumped Milton Friedman except that we increase both
We set the cost
We set the probability distributions
Below is some Python code that sets up these objects.
@jit
def p(x, a, b):
"Beta distribution."
r = gamma(a + b) / (gamma(a) * gamma(b))
return r * x**(a-1) * (1 - x)**(b-1)
We start with defining a jitclass that stores parameters and
functions we need to solve problems for both the Bayesian and
frequentist Navy Captains.
wf_data = [
('c', float64), # unemployment compensation
('a0', float64), # parameters of beta distribution
('b0', float64),
('a1', float64),
('b1', float64),
('L0', float64), # cost of selecting f0 when f1 is true
('L1', float64), # cost of selecting f1 when f0 is true
('π_grid', float64[:]), # grid of beliefs π
('π_grid_size', int64),
('mc_size', int64), # size of Monto Carlo simulation
('z0', float64[:]), # sequence of random values
('z1', float64[:]) # sequence of random values
]
@jitclass(wf_data)
class WaldFriedman:
def __init__(self,
c=1.25,
a0=1,
b0=1,
a1=3,
b1=1.2,
L0=100,
L1=100,
π_grid_size=200,
mc_size=1000):
self.c, self.π_grid_size = c, π_grid_size
self.a0, self.b0, self.a1, self.b1 = a0, b0, a1, b1
self.L0, self.L1 = L0, L1
self.π_grid = np.linspace(0, 1, π_grid_size)
self.mc_size = mc_size
self.z0 = np.random.beta(a0, b0, mc_size)
self.z1 = np.random.beta(a1, b1, mc_size)
def f0(self, x):
return p(x, self.a0, self.b0)
def f1(self, x):
return p(x, self.a1, self.b1)
def κ(self, z, π):
"""
Updates π using Bayes' rule and the current observation z
"""
a0, b0, a1, b1 = self.a0, self.b0, self.a1, self.b1
π_f0, π_f1 = π * p(z, a0, b0), (1 - π) * p(z, a1, b1)
π_new = π_f0 / (π_f0 + π_f1)
return π_new
wf = WaldFriedman()
grid = np.linspace(0, 1, 50)
plt.figure()
plt.title("Two Distributions")
plt.plot(grid, wf.f0(grid), lw=2, label="$f_0$")
plt.plot(grid, wf.f1(grid), lw=2, label="$f_1$")
plt.legend()
plt.xlabel("$z$ values")
plt.ylabel("density of $z_k$")
plt.tight_layout()
plt.show()
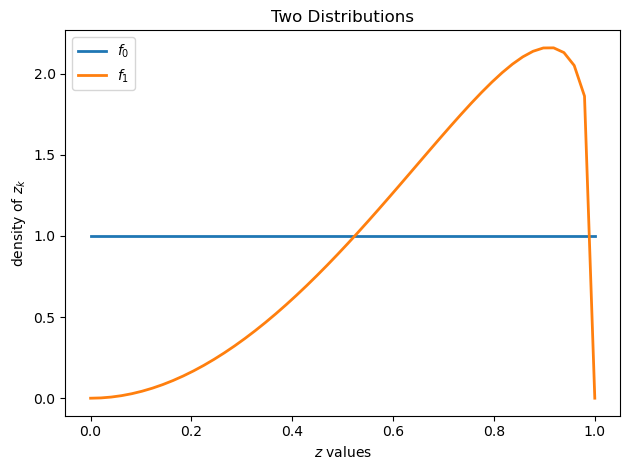
Above, we plot the two possible probability densities
56.3. Frequentist Decision Rule#
The Navy told the Captain to use a frequentist decision rule.
In particular, it gave him a decision rule that the Navy had designed by using frequentist statistical theory to minimize an expected loss function.
That decision rule is characterized by
a sample size
a cutoff value
Let
The decision rule associated with a sample size
decide that
decide that
To understand how that rule was engineered, let null and alternative hypotheses be
null:
alternative
Given sample size
Here
For a given sample size
by choosing
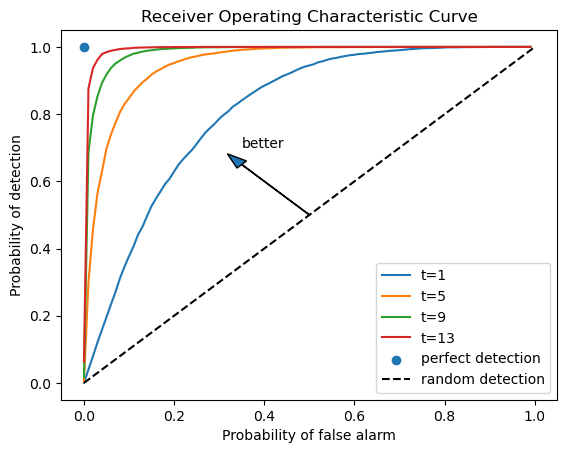
To see some receiver operating characteristic curves, please see this lecture Likelihood Ratio Processes.
To solve for
N = 10000
T = 100
z0_arr = np.random.beta(wf.a0, wf.b0, (N, T))
z1_arr = np.random.beta(wf.a1, wf.b1, (N, T))
plt.hist(z0_arr.flatten(), bins=50, alpha=0.4, label='f0')
plt.hist(z1_arr.flatten(), bins=50, alpha=0.4, label='f1')
plt.legend()
plt.show()
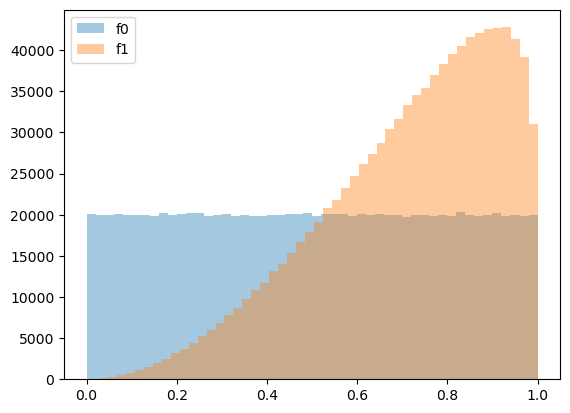
We can compute sequences of likelihood ratios using simulated samples.
l = lambda z: wf.f0(z) / wf.f1(z)
l0_arr = l(z0_arr)
l1_arr = l(z1_arr)
L0_arr = np.cumprod(l0_arr, 1)
L1_arr = np.cumprod(l1_arr, 1)
With an empirical distribution of likelihood ratios in hand, we can draw
receiver operating characteristic curves by enumerating
PFA = np.arange(0, 100, 1)
for t in range(1, 15, 4):
percentile = np.percentile(L0_arr[:, t], PFA)
PD = [np.sum(L1_arr[:, t] < p) / N for p in percentile]
plt.plot(PFA / 100, PD, label=f"t={t}")
plt.scatter(0, 1, label="perfect detection")
plt.plot([0, 1], [0, 1], color='k', ls='--', label="random detection")
plt.arrow(0.5, 0.5, -0.15, 0.15, head_width=0.03)
plt.text(0.35, 0.7, "better")
plt.xlabel("Probability of false alarm")
plt.ylabel("Probability of detection")
plt.legend()
plt.title("Receiver Operating Characteristic Curve")
plt.show()
Our frequentist minimizes the expected total loss presented in equation
(56.1) by choosing
Doing that delivers an expected loss
We first consider the case in which
We can solve the minimization problem in two steps.
First, we fix
Here is Python code that does that and then plots a useful graph.
@jit
def V_fre_d_t(d, t, L0_arr, L1_arr, π_star, wf):
N = L0_arr.shape[0]
PFA = np.sum(L0_arr[:, t-1] < d) / N
PD = np.sum(L1_arr[:, t-1] < d) / N
V = π_star * PFA *wf. L1 + (1 - π_star) * (1 - PD) * wf.L0
return V
def V_fre_t(t, L0_arr, L1_arr, π_star, wf):
res = minimize(V_fre_d_t, 1, args=(t, L0_arr, L1_arr, π_star, wf), method='Nelder-Mead')
V = res.fun
d = res.x
PFA = np.sum(L0_arr[:, t-1] < d) / N
PD = np.sum(L1_arr[:, t-1] < d) / N
return V, PFA, PD
def compute_V_fre(L0_arr, L1_arr, π_star, wf):
T = L0_arr.shape[1]
V_fre_arr = np.empty(T)
PFA_arr = np.empty(T)
PD_arr = np.empty(T)
for t in range(1, T+1):
V, PFA, PD = V_fre_t(t, L0_arr, L1_arr, π_star, wf)
V_fre_arr[t-1] = wf.c * t + V
PFA_arr[t-1] = PFA
PD_arr[t-1] = PD
return V_fre_arr, PFA_arr, PD_arr
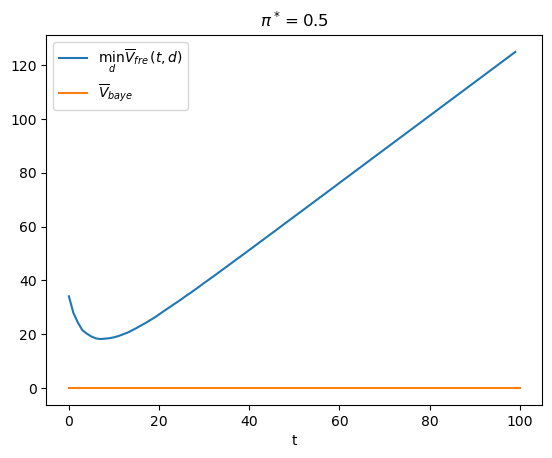
π_star = 0.5
V_fre_arr, PFA_arr, PD_arr = compute_V_fre(L0_arr, L1_arr, π_star, wf)
plt.plot(range(T), V_fre_arr, label=r'$\min_{d} \overline{V}_{fre}(t,d)$')
plt.xlabel('t')
plt.title(r'$\pi^*=0.5$')
plt.legend()
plt.show()
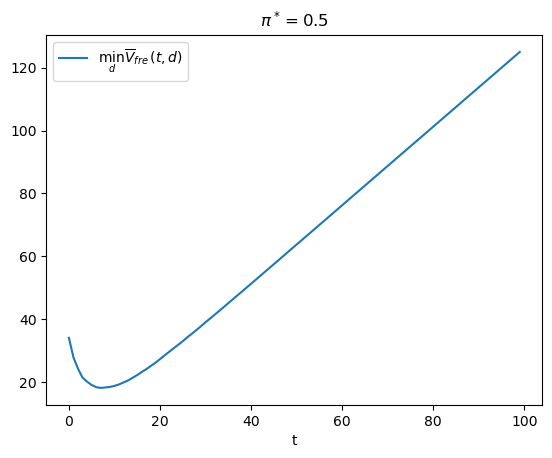
t_optimal = np.argmin(V_fre_arr) + 1
msg = f"The above graph indicates that minimizing over t tells the frequentist to draw {t_optimal} observations and then decide."
print(msg)
The above graph indicates that minimizing over t tells the frequentist to draw 8 observations and then decide.
Let’s now change the value of
n_π = 20
π_star_arr = np.linspace(0.1, 0.9, n_π)
V_fre_bar_arr = np.empty(n_π)
t_optimal_arr = np.empty(n_π)
PFA_optimal_arr = np.empty(n_π)
PD_optimal_arr = np.empty(n_π)
for i, π_star in enumerate(π_star_arr):
V_fre_arr, PFA_arr, PD_arr = compute_V_fre(L0_arr, L1_arr, π_star, wf)
t_idx = np.argmin(V_fre_arr)
V_fre_bar_arr[i] = V_fre_arr[t_idx]
t_optimal_arr[i] = t_idx + 1
PFA_optimal_arr[i] = PFA_arr[t_idx]
PD_optimal_arr[i] = PD_arr[t_idx]
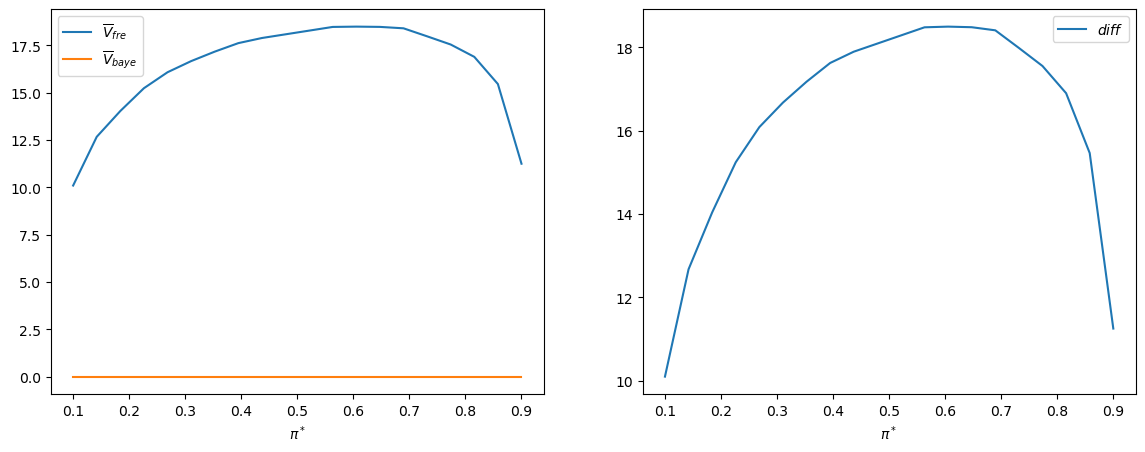
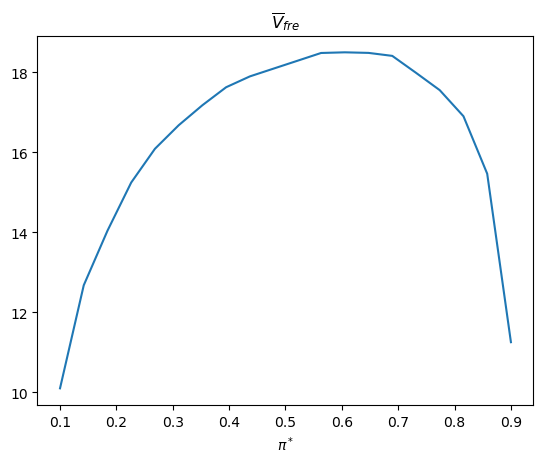
plt.plot(π_star_arr, V_fre_bar_arr)
plt.xlabel(r'$\pi^*$')
plt.title(r'$\overline{V}_{fre}$')
plt.show()
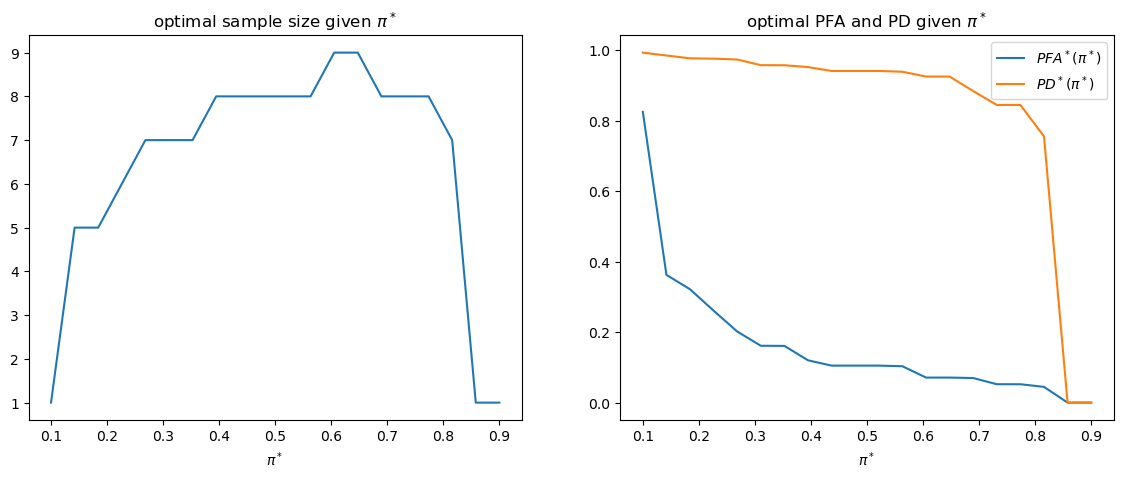
The following shows how optimal sample size
fig, axs = plt.subplots(1, 2, figsize=(14, 5))
axs[0].plot(π_star_arr, t_optimal_arr)
axs[0].set_xlabel(r'$\pi^*$')
axs[0].set_title('optimal sample size given $\pi^*$')
axs[1].plot(π_star_arr, PFA_optimal_arr, label='$PFA^*(\pi^*)$')
axs[1].plot(π_star_arr, PD_optimal_arr, label='$PD^*(\pi^*)$')
axs[1].set_xlabel(r'$\pi^*$')
axs[1].legend()
axs[1].set_title('optimal PFA and PD given $\pi^*$')
plt.show()
<>:5: SyntaxWarning: invalid escape sequence '\p'
<>:7: SyntaxWarning: invalid escape sequence '\p'
<>:8: SyntaxWarning: invalid escape sequence '\p'
<>:11: SyntaxWarning: invalid escape sequence '\p'
<>:5: SyntaxWarning: invalid escape sequence '\p'
<>:7: SyntaxWarning: invalid escape sequence '\p'
<>:8: SyntaxWarning: invalid escape sequence '\p'
<>:11: SyntaxWarning: invalid escape sequence '\p'
/tmp/ipykernel_8110/677325419.py:5: SyntaxWarning: invalid escape sequence '\p'
axs[0].set_title('optimal sample size given $\pi^*$')
/tmp/ipykernel_8110/677325419.py:7: SyntaxWarning: invalid escape sequence '\p'
axs[1].plot(π_star_arr, PFA_optimal_arr, label='$PFA^*(\pi^*)$')
/tmp/ipykernel_8110/677325419.py:8: SyntaxWarning: invalid escape sequence '\p'
axs[1].plot(π_star_arr, PD_optimal_arr, label='$PD^*(\pi^*)$')
/tmp/ipykernel_8110/677325419.py:11: SyntaxWarning: invalid escape sequence '\p'
axs[1].set_title('optimal PFA and PD given $\pi^*$')
56.4. Bayesian Decision Rule#
In A Problem that Stumped Milton Friedman, we learned how Abraham Wald confirmed the Navy Captain’s hunch that there is a better decision rule.
We presented a Bayesian procedure that instructed the Captain to makes
decisions by comparing his current Bayesian posterior probability
To proceed, we borrow some Python code from the quantecon
lecture A Problem that Stumped Milton Friedman
that computes
@jit(parallel=True)
def Q(h, wf):
c, π_grid = wf.c, wf.π_grid
L0, L1 = wf.L0, wf.L1
z0, z1 = wf.z0, wf.z1
mc_size = wf.mc_size
κ = wf.κ
h_new = np.empty_like(π_grid)
h_func = lambda p: np.interp(p, π_grid, h)
for i in prange(len(π_grid)):
π = π_grid[i]
# Find the expected value of J by integrating over z
integral_f0, integral_f1 = 0, 0
for m in range(mc_size):
π_0 = κ(z0[m], π) # Draw z from f0 and update π
integral_f0 += min((1 - π_0) * L0, π_0 * L1, h_func(π_0))
π_1 = κ(z1[m], π) # Draw z from f1 and update π
integral_f1 += min((1 - π_1) * L0, π_1 * L1, h_func(π_1))
integral = (π * integral_f0 + (1 - π) * integral_f1) / mc_size
h_new[i] = c + integral
return h_new
@jit
def solve_model(wf, tol=1e-4, max_iter=1000):
"""
Compute the continuation value function
* wf is an instance of WaldFriedman
"""
# Set up loop
h = np.zeros(len(wf.π_grid))
i = 0
error = tol + 1
while i < max_iter and error > tol:
h_new = Q(h, wf)
error = np.max(np.abs(h - h_new))
i += 1
h = h_new
if error > tol:
print("Failed to converge!")
return h_new
h_star = solve_model(wf)
@jit
def find_cutoff_rule(wf, h):
"""
This function takes a continuation value function and returns the
corresponding cutoffs of where you transition between continuing and
choosing a specific model
"""
π_grid = wf.π_grid
L0, L1 = wf.L0, wf.L1
# Evaluate cost at all points on grid for choosing a model
payoff_f0 = (1 - π_grid) * L0
payoff_f1 = π_grid * L1
# The cutoff points can be found by differencing these costs with
# The Bellman equation (J is always less than or equal to p_c_i)
β = π_grid[np.searchsorted(
payoff_f1 - np.minimum(h, payoff_f0),
1e-10)
- 1]
α = π_grid[np.searchsorted(
np.minimum(h, payoff_f1) - payoff_f0,
1e-10)
- 1]
return (β, α)
β, α = find_cutoff_rule(wf, h_star)
cost_L0 = (1 - wf.π_grid) * wf.L0
cost_L1 = wf.π_grid * wf.L1
fig, ax = plt.subplots(figsize=(10, 6))
ax.plot(wf.π_grid, h_star, label='continuation value')
ax.plot(wf.π_grid, cost_L1, label='choose f1')
ax.plot(wf.π_grid, cost_L0, label='choose f0')
ax.plot(wf.π_grid,
np.amin(np.column_stack([h_star, cost_L0, cost_L1]),axis=1),
lw=15, alpha=0.1, color='b', label='minimum cost')
ax.annotate(r"$\beta$", xy=(β + 0.01, 0.5), fontsize=14)
ax.annotate(r"$\alpha$", xy=(α + 0.01, 0.5), fontsize=14)
plt.vlines(β, 0, β * wf.L0, linestyle="--")
plt.vlines(α, 0, (1 - α) * wf.L1, linestyle="--")
ax.set(xlim=(0, 1), ylim=(0, 0.5 * max(wf.L0, wf.L1)), ylabel="cost",
xlabel=r"$\pi$", title="Value function")
plt.legend(borderpad=1.1)
plt.show()
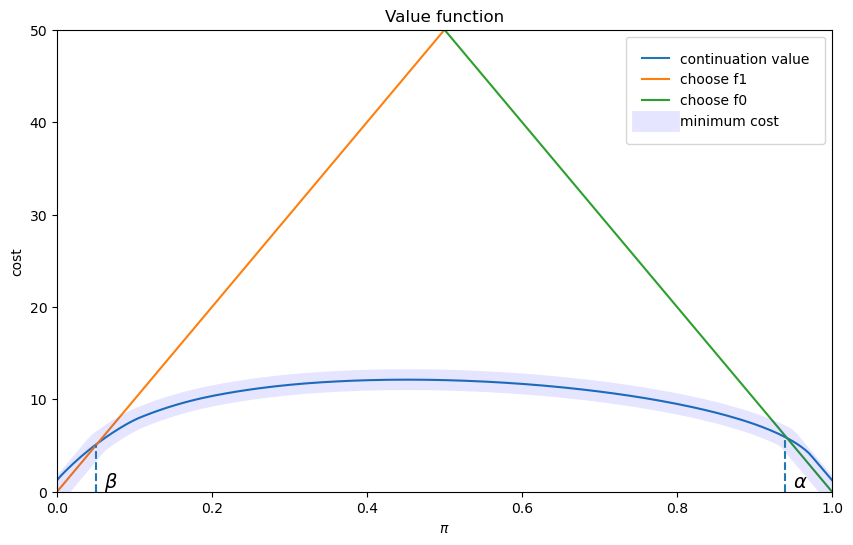
The above figure portrays the value function plotted against the decision maker’s Bayesian posterior.
It also shows the probabilities
The Bayesian decision rule is:
accept
accept
delay deciding and draw another
We can calculate two “objective” loss functions under this situation
conditioning on knowing for sure that nature has selected
under
under
where
Given a prior probability
Below we write some Python code that computes
@jit(parallel=True)
def V_q(wf, flag):
V = np.zeros(wf.π_grid_size)
if flag == 0:
z_arr = wf.z0
V[wf.π_grid < β] = wf.L1
else:
z_arr = wf.z1
V[wf.π_grid >= α] = wf.L0
V_old = np.empty_like(V)
while True:
V_old[:] = V[:]
V[(β <= wf.π_grid) & (wf.π_grid < α)] = 0
for i in prange(len(wf.π_grid)):
π = wf.π_grid[i]
if π >= α or π < β:
continue
for j in prange(len(z_arr)):
π_next = wf.κ(z_arr[j], π)
V[i] += wf.c + np.interp(π_next, wf.π_grid, V_old)
V[i] /= wf.mc_size
if np.abs(V - V_old).max() < 1e-5:
break
return V
V0 = V_q(wf, 0)
V1 = V_q(wf, 1)
plt.plot(wf.π_grid, V0, label='$V^0$')
plt.plot(wf.π_grid, V1, label='$V^1$')
plt.vlines(β, 0, wf.L0, linestyle='--')
plt.text(β+0.01, wf.L0/2, 'β')
plt.vlines(α, 0, wf.L0, linestyle='--')
plt.text(α+0.01, wf.L0/2, 'α')
plt.xlabel(r'$\pi$')
plt.title('Objective value function $V(\pi)$')
plt.legend()
plt.show()
<>:11: SyntaxWarning: invalid escape sequence '\p'
<>:11: SyntaxWarning: invalid escape sequence '\p'
/tmp/ipykernel_8110/4049178433.py:11: SyntaxWarning: invalid escape sequence '\p'
plt.title('Objective value function $V(\pi)$')
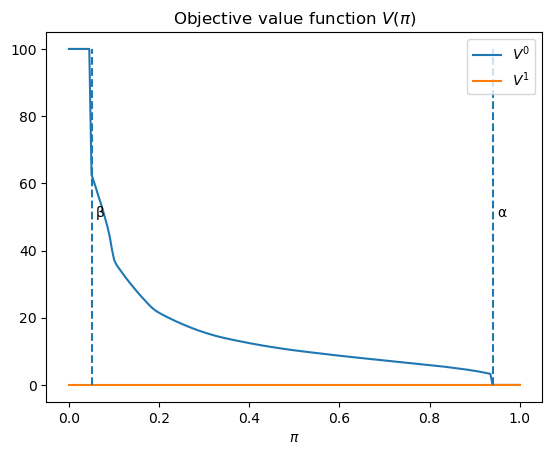
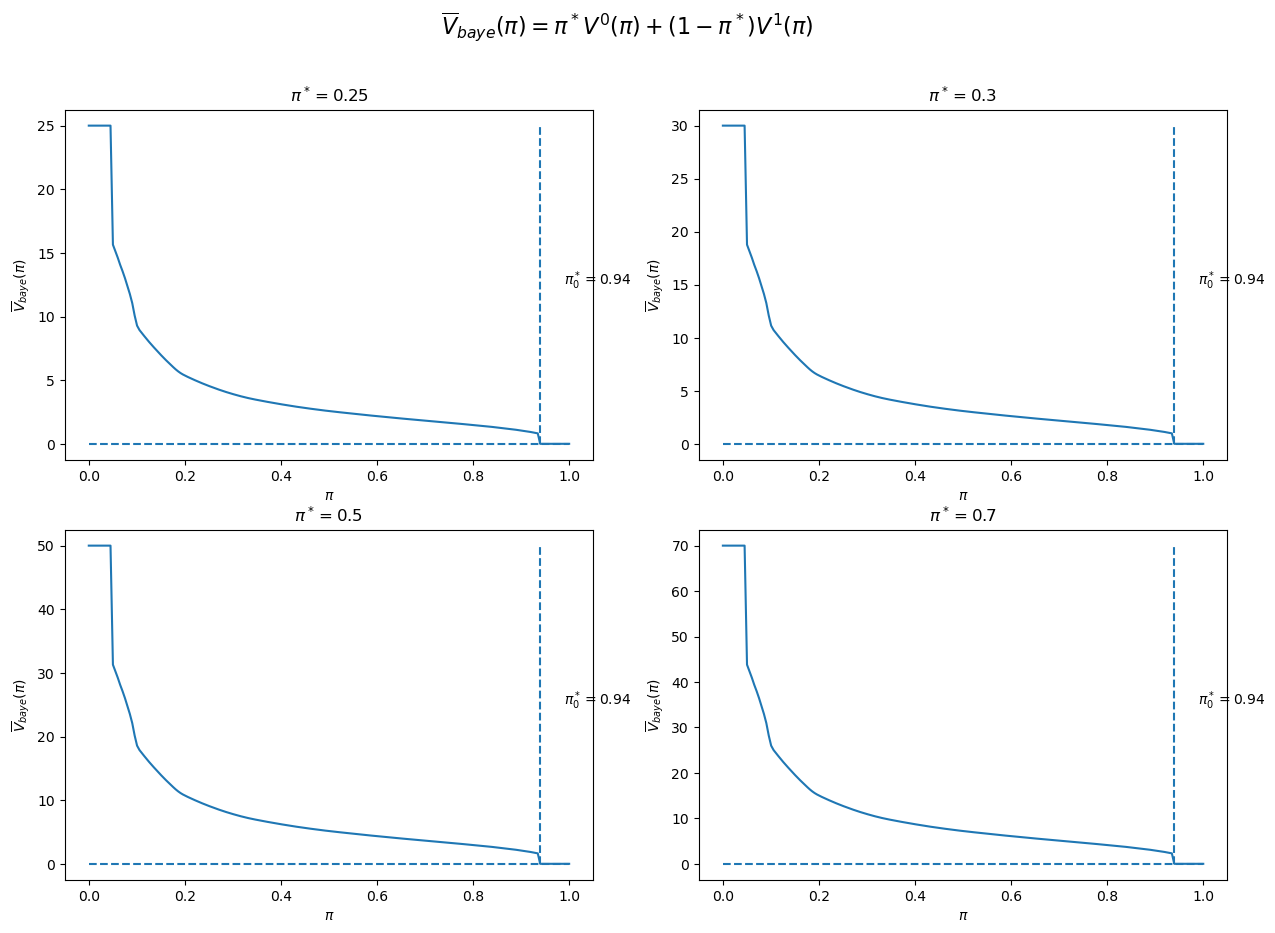
Given an assumed value for
We can then determine an initial Bayesian prior
The figure below plots four cases corresponding to
We observe that in each case
def compute_V_baye_bar(π_star, V0, V1, wf):
V_baye = π_star * V0 + (1 - π_star) * V1
π_idx = np.argmin(V_baye)
π_optimal = wf.π_grid[π_idx]
V_baye_bar = V_baye[π_idx]
return V_baye, π_optimal, V_baye_bar
π_star_arr = [0.25, 0.3, 0.5, 0.7]
fig, axs = plt.subplots(2, 2, figsize=(15, 10))
for i, π_star in enumerate(π_star_arr):
row_i = i // 2
col_i = i % 2
V_baye, π_optimal, V_baye_bar = compute_V_baye_bar(π_star, V0, V1, wf)
axs[row_i, col_i].plot(wf.π_grid, V_baye)
axs[row_i, col_i].hlines(V_baye_bar, 0, 1, linestyle='--')
axs[row_i, col_i].vlines(π_optimal, V_baye_bar, V_baye.max(), linestyle='--')
axs[row_i, col_i].text(π_optimal+0.05, (V_baye_bar + V_baye.max()) / 2,
'${\pi_0^*}=$'+f'{π_optimal:0.2f}')
axs[row_i, col_i].set_xlabel(r'$\pi$')
axs[row_i, col_i].set_ylabel(r'$\overline{V}_{baye}(\pi)$')
axs[row_i, col_i].set_title(r'$\pi^*=$' + f'{π_star}')
fig.suptitle(r'$\overline{V}_{baye}(\pi)=\pi^*V^0(\pi) + (1-\pi^*)V^1(\pi)$', fontsize=16)
plt.show()
<>:15: SyntaxWarning: invalid escape sequence '\p'
<>:15: SyntaxWarning: invalid escape sequence '\p'
/tmp/ipykernel_8110/3947394423.py:15: SyntaxWarning: invalid escape sequence '\p'
'${\pi_0^*}=$'+f'{π_optimal:0.2f}')
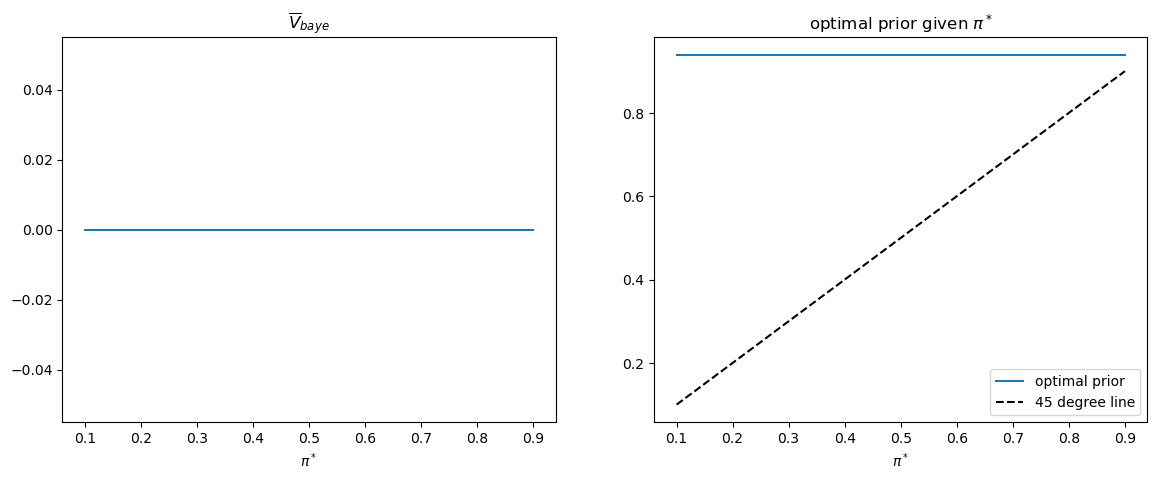
This pattern of outcomes holds more generally.
Thus, the following Python code generates the associated graph that
verifies the equality of
π_star_arr = np.linspace(0.1, 0.9, n_π)
V_baye_bar_arr = np.empty_like(π_star_arr)
π_optimal_arr = np.empty_like(π_star_arr)
for i, π_star in enumerate(π_star_arr):
V_baye, π_optimal, V_baye_bar = compute_V_baye_bar(π_star, V0, V1, wf)
V_baye_bar_arr[i] = V_baye_bar
π_optimal_arr[i] = π_optimal
fig, axs = plt.subplots(1, 2, figsize=(14, 5))
axs[0].plot(π_star_arr, V_baye_bar_arr)
axs[0].set_xlabel(r'$\pi^*$')
axs[0].set_title(r'$\overline{V}_{baye}$')
axs[1].plot(π_star_arr, π_optimal_arr, label='optimal prior')
axs[1].plot([π_star_arr.min(), π_star_arr.max()],
[π_star_arr.min(), π_star_arr.max()],
c='k', linestyle='--', label='45 degree line')
axs[1].set_xlabel(r'$\pi^*$')
axs[1].set_title('optimal prior given $\pi^*$')
axs[1].legend()
plt.show()
<>:23: SyntaxWarning: invalid escape sequence '\p'
<>:23: SyntaxWarning: invalid escape sequence '\p'
/tmp/ipykernel_8110/90043130.py:23: SyntaxWarning: invalid escape sequence '\p'
axs[1].set_title('optimal prior given $\pi^*$')
56.6. More Details#
We can provide more insights by focusing on the case in which
π_star = 0.5
Recall that when t_optimal ex ante.
For our parameter settings, we can compute its value:
t_optimal
8
For convenience, let’s define t_idx as the Python array index
corresponding to t_optimal sample size.
t_idx = t_optimal - 1
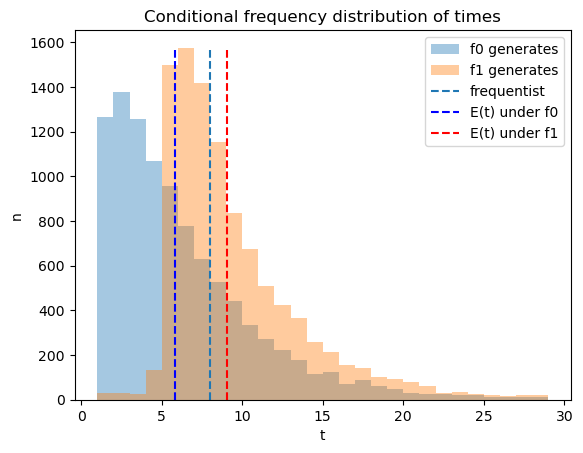
56.7. Distribution of Bayesian Decision Rule’s Time to Decide#
We use simulations to compute the frequency distribution of the time to
decide for the Bayesian decision rule and compare that time to the
frequentist rule’s fixed
The following Python code creates a graph that shows the frequency
distribution of Bayesian times to decide of Bayesian decision maker,
conditional on distribution
The blue and red dotted lines show averages for the Bayesian decision
rule, while the black dotted line shows the frequentist optimal sample
size
On average the Bayesian rule decides earlier than the frequentist
rule when
@jit(parallel=True)
def check_results(L_arr, α, β, flag, π0):
N, T = L_arr.shape
time_arr = np.empty(N)
correctness = np.empty(N)
π_arr = π0 * L_arr / (π0 * L_arr + 1 - π0)
for i in prange(N):
for t in range(T):
if (π_arr[i, t] < β) or (π_arr[i, t] > α):
time_arr[i] = t + 1
correctness[i] = (flag == 0 and π_arr[i, t] > α) or (flag == 1 and π_arr[i, t] < β)
break
return time_arr, correctness
time_arr0, correctness0 = check_results(L0_arr, α, β, 0, π_star)
time_arr1, correctness1 = check_results(L1_arr, α, β, 1, π_star)
# unconditional distribution
time_arr_u = np.concatenate((time_arr0, time_arr1))
correctness_u = np.concatenate((correctness0, correctness1))
n1 = plt.hist(time_arr0, bins=range(1, 30), alpha=0.4, label='f0 generates')[0]
n2 = plt.hist(time_arr1, bins=range(1, 30), alpha=0.4, label='f1 generates')[0]
plt.vlines(t_optimal, 0, max(n1.max(), n2.max()), linestyle='--', label='frequentist')
plt.vlines(np.mean(time_arr0), 0, max(n1.max(), n2.max()),
linestyle='--', color='b', label='E(t) under f0')
plt.vlines(np.mean(time_arr1), 0, max(n1.max(), n2.max()),
linestyle='--', color='r', label='E(t) under f1')
plt.legend();
plt.xlabel('t')
plt.ylabel('n')
plt.title('Conditional frequency distribution of times')
plt.show()
Later we’ll figure out how these distributions ultimately affect objective expected values under the two decision rules.
To begin, let’s look at simulations of the Bayesian’s beliefs over time.
We can easily compute the updated beliefs at any time
π0_arr = π_star * L0_arr / (π_star * L0_arr + 1 - π_star)
π1_arr = π_star * L1_arr / (π_star * L1_arr + 1 - π_star)
fig, axs = plt.subplots(1, 2, figsize=(14, 4))
axs[0].plot(np.arange(1, π0_arr.shape[1]+1), np.mean(π0_arr, 0), label='f0 generates')
axs[0].plot(np.arange(1, π1_arr.shape[1]+1), 1 - np.mean(π1_arr, 0), label='f1 generates')
axs[0].set_xlabel('t')
axs[0].set_ylabel('$E(\pi_t)$ or ($1 - E(\pi_t)$)')
axs[0].set_title('Expectation of beliefs after drawing t observations')
axs[0].legend()
axs[1].plot(np.arange(1, π0_arr.shape[1]+1), np.var(π0_arr, 0), label='f0 generates')
axs[1].plot(np.arange(1, π1_arr.shape[1]+1), np.var(π1_arr, 0), label='f1 generates')
axs[1].set_xlabel('t')
axs[1].set_ylabel('var($\pi_t$)')
axs[1].set_title('Variance of beliefs after drawing t observations')
axs[1].legend()
plt.show()
<>:6: SyntaxWarning: invalid escape sequence '\p'
<>:13: SyntaxWarning: invalid escape sequence '\p'
<>:6: SyntaxWarning: invalid escape sequence '\p'
<>:13: SyntaxWarning: invalid escape sequence '\p'
/tmp/ipykernel_8110/2416490984.py:6: SyntaxWarning: invalid escape sequence '\p'
axs[0].set_ylabel('$E(\pi_t)$ or ($1 - E(\pi_t)$)')
/tmp/ipykernel_8110/2416490984.py:13: SyntaxWarning: invalid escape sequence '\p'
axs[1].set_ylabel('var($\pi_t$)')
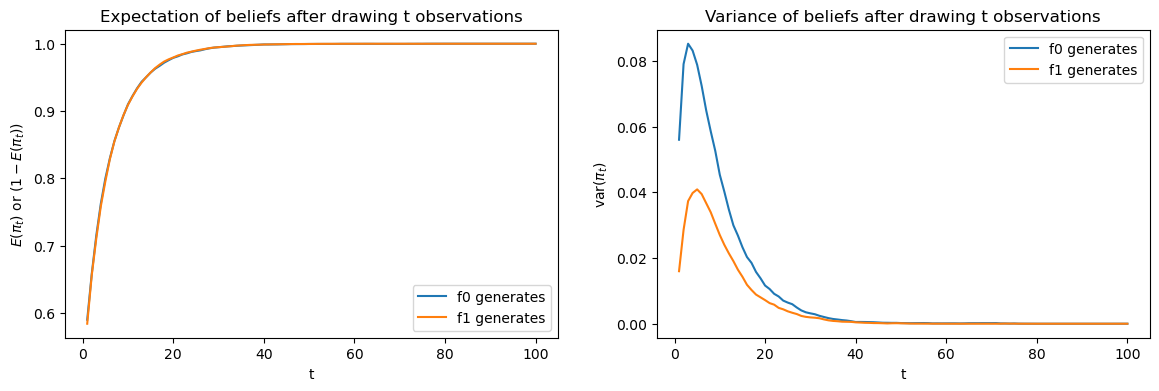
The above figures compare averages and variances of updated Bayesian
posteriors after
The left graph compares
However, as the right hand size graph shows, there is significant
difference in variances when
The difference in variances is the reason that the Bayesian decision
maker waits longer to decide when
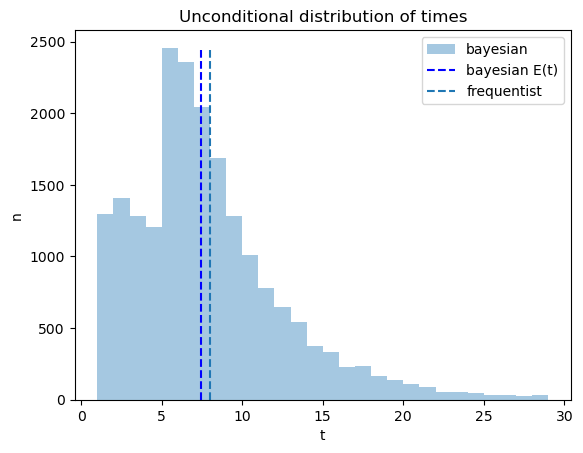
The code below plots outcomes of constructing an unconditional
distribution by simply pooling the simulated data across the two
possible distributions
The pooled distribution describes a sense in which on average the Bayesian decides earlier, an outcome that seems at least partly to confirm the Navy Captain’s hunch.
n = plt.hist(time_arr_u, bins=range(1, 30), alpha=0.4, label='bayesian')[0]
plt.vlines(np.mean(time_arr_u), 0, n.max(), linestyle='--',
color='b', label='bayesian E(t)')
plt.vlines(t_optimal, 0, n.max(), linestyle='--', label='frequentist')
plt.legend()
plt.xlabel('t')
plt.ylabel('n')
plt.title('Unconditional distribution of times')
plt.show()
56.8. Probability of Making Correct Decision#
Now we use simulations to compute the fraction of samples in which the Bayesian and the frequentist decision rules decide correctly.
For the frequentist rule, the probability of making the correct decision
under
Below we plot these two probabilities for the frequentist rule, along
with the conditional probabilities that the Bayesian rule decides before
# optimal PFA and PD of frequentist with optimal sample size
V, PFA, PD = V_fre_t(t_optimal, L0_arr, L1_arr, π_star, wf)
plt.plot([1, 20], [PD, PD], linestyle='--', label='PD: fre. chooses f1 correctly')
plt.plot([1, 20], [1-PFA, 1-PFA], linestyle='--', label='1-PFA: fre. chooses f0 correctly')
plt.vlines(t_optimal, 0, 1, linestyle='--', label='frequentist optimal sample size')
N = time_arr0.size
T_arr = np.arange(1, 21)
plt.plot(T_arr, [np.sum(correctness0[time_arr0 <= t] == 1) / N for t in T_arr],
label='q=f0 and baye. choose f0')
plt.plot(T_arr, [np.sum(correctness1[time_arr1 <= t] == 1) / N for t in T_arr],
label='q=f1 and baye. choose f1')
plt.legend(loc=4)
plt.xlabel('t')
plt.ylabel('Probability')
plt.title('Cond. probability of making correct decisions before t')
plt.show()
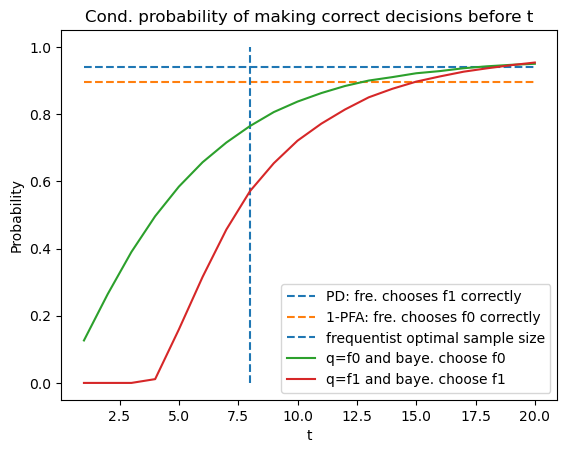
By averaging using
plt.plot([1, 20], [(PD + 1 - PFA) / 2, (PD + 1 - PFA) / 2],
linestyle='--', label='fre. makes correct decision')
plt.vlines(t_optimal, 0, 1, linestyle='--', label='frequentist optimal sample size')
N = time_arr_u.size
plt.plot(T_arr, [np.sum(correctness_u[time_arr_u <= t] == 1) / N for t in T_arr],
label="bayesian makes correct decision")
plt.legend()
plt.xlabel('t')
plt.ylabel('Probability')
plt.title('Uncond. probability of making correct decisions before t')
plt.show()
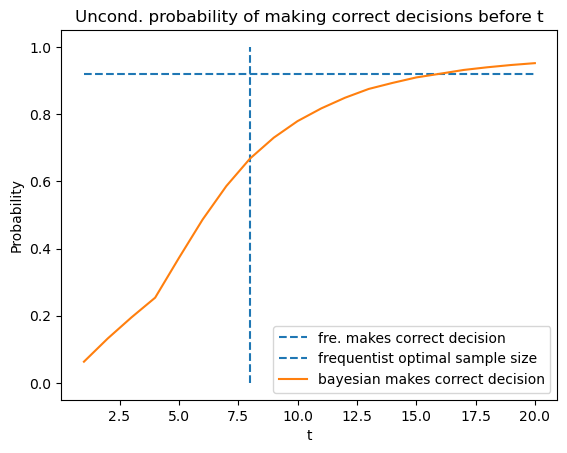
56.9. Distribution of Likelihood Ratios at Frequentist’s
Next we use simulations to construct distributions of likelihood ratios
after
To serve as useful reference points, we also show likelihood ratios that
correspond to the Bayesian cutoffs
In order to exhibit the distribution more clearly, we report logarithms of likelihood ratios.
The graphs below reports two distributions, one conditional on
Lα = (1 - π_star) * α / (π_star - π_star * α)
Lβ = (1 - π_star) * β / (π_star - π_star * β)
L_min = min(L0_arr[:, t_idx].min(), L1_arr[:, t_idx].min())
L_max = max(L0_arr[:, t_idx].max(), L1_arr[:, t_idx].max())
bin_range = np.linspace(np.log(L_min), np.log(L_max), 50)
n0 = plt.hist(np.log(L0_arr[:, t_idx]), bins=bin_range, alpha=0.4, label='f0 generates')[0]
n1 = plt.hist(np.log(L1_arr[:, t_idx]), bins=bin_range, alpha=0.4, label='f1 generates')[0]
plt.vlines(np.log(Lβ), 0, max(n0.max(), n1.max()), linestyle='--', color='r', label='log($L_β$)')
plt.vlines(np.log(Lα), 0, max(n0.max(), n1.max()), linestyle='--', color='b', label='log($L_α$)')
plt.legend()
plt.xlabel('log(L)')
plt.ylabel('n')
plt.title('Cond. distribution of log likelihood ratio at frequentist t')
plt.show()
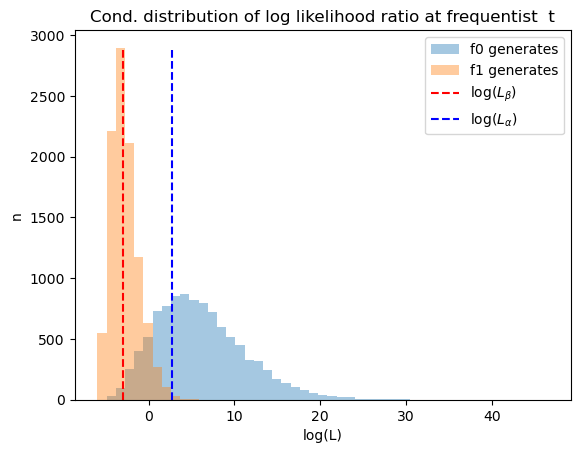
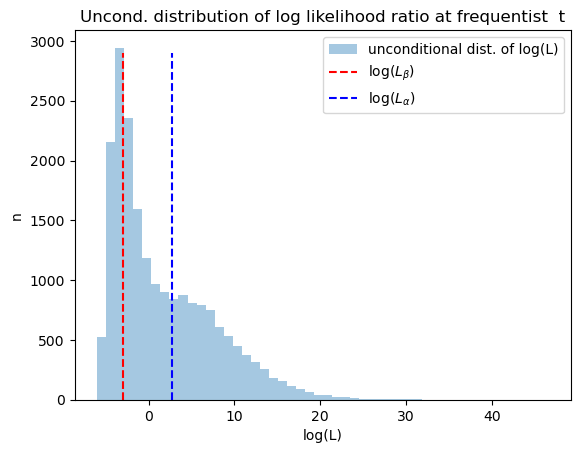
The next graph plots the unconditional distribution of Bayesian times to decide, constructed as earlier by pooling the two conditional distributions.
plt.hist(np.log(np.concatenate([L0_arr[:, t_idx], L1_arr[:, t_idx]])),
bins=50, alpha=0.4, label='unconditional dist. of log(L)')
plt.vlines(np.log(Lβ), 0, max(n0.max(), n1.max()), linestyle='--', color='r', label='log($L_β$)')
plt.vlines(np.log(Lα), 0, max(n0.max(), n1.max()), linestyle='--', color='b', label='log($L_α$)')
plt.legend()
plt.xlabel('log(L)')
plt.ylabel('n')
plt.title('Uncond. distribution of log likelihood ratio at frequentist t')
plt.show()